PocketDepth - A Depth Based Algorithm for Identification of Ligand Binding Sites
About PocketDepth
Computational methods for identifying and predicting functional sites in protein structures are crucial in structural biology and bioinformatics. These methods not only enhance our understanding of molecular function but also contribute to structure-based design of ligands, drugs, and modified protein molecules. Despite existing structure-based prediction methods, the complexity and diversity of protein structures demand the exploration of newer methods. This abstract introduces the PocketDepth method, a novel approach for identifying binding sites in proteins.
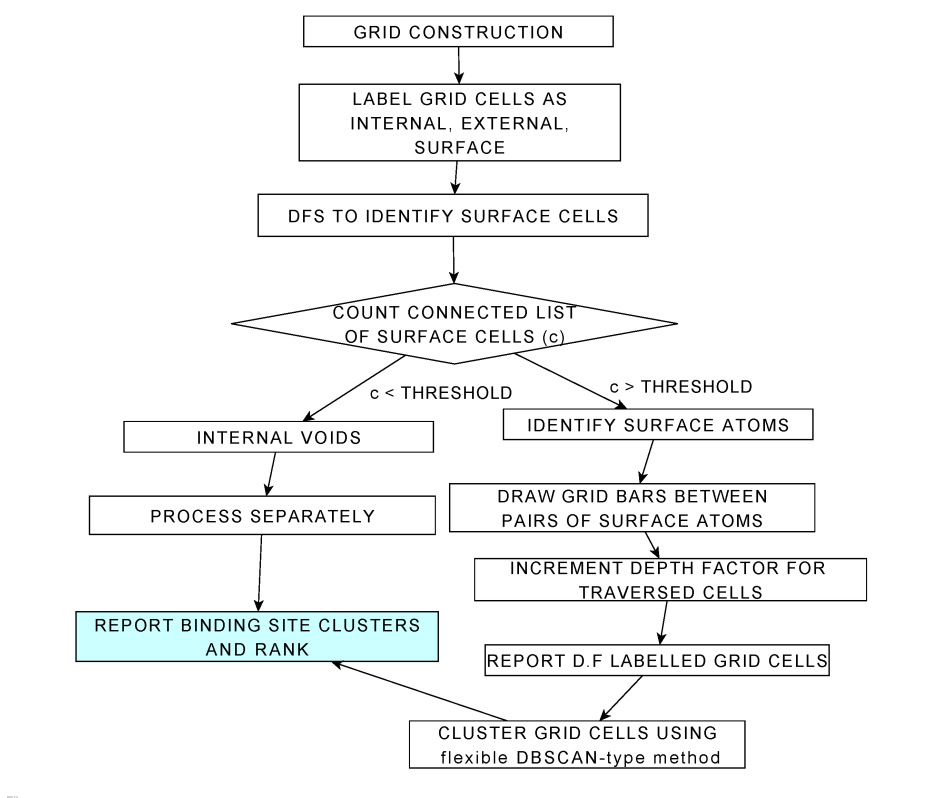
PocketDepth is a geometry-based method that operates in two stages: labeling grid cells with depth factors and depth-based clustering using neighborhood information. Depth, a significant parameter in protein structure analysis, is systematically considered in our method. It reflects the centrality of a sub-space to a putative pocket rather than merely indicating its distance from the protein's external surface. The algorithm's performance was evaluated against PDBbind, a curated set of 1091 proteins, achieving a prediction accuracy of approximately 96%. Notably, 87% of true-positives were identified within the top five ranks, with 55% in the first rank itself. Furthermore, 77% of predictions had at least 50% overlap with experimentally observed ligands.
Extensive testing against a dataset of apo-proteins and their respective ligand complexes demonstrated high prediction rates. A comparative analysis with four widely used methods on a representative set is also presented. The PocketDepth method showcases its accuracy, versatility, and efficiency in predicting binding sites in diverse protein structures.
Algorithm
Flow chart depicting the basic idea behind the algorithm: